Chapter 8: Beyond Standard Regression Analysis
Chapter08.RmdSection 8.1: Binary response variables
Example 8.1: Labor market data
We illustrate probit regression analysis for the labor market data.
library("BayesianLearningCode")
data("labor", package = "BayesianLearningCode")We model the binary variable unemployment as dependent variable and use as covariates the variables female (binary), wcollar (binary), age18 (quantitative, centered at 18 years) and unemployed in 1997 (binary). The baseline person is hence an 18 year old male blue collar worker who was employed in 1997.
y.unemp <- labor$income_1998 == "zero"
N.unemp <- length(y.unemp) # number of observations
X.unemp <- with(labor, cbind(intercept = rep(1, N.unemp),
female = female,
age18 = 1998 - birthyear - 18,
wcollar = wcollar_1997,
unemp97 = income_1997 == "zero")) # regressor matrixThe regression coefficients are estimated using data augmentation and Gibbs sampling. We define a function yielding posterior draws using the algorithm detailed in Chapter 8.1.1.
probit <- function(y, X, b0 = 0, B0 = 10000,
burnin = 1000L, M = 5000L) {
N <- length(y)
d <- ncol(X) # number regression effects
B0.inv <- diag(rep(1 / B0, length.out = d), nrow = d)
b0 <- rep(b0, length.out = d)
B0inv.b0 <- B0.inv %*% b0
betas <- matrix(NA_real_, nrow = M, ncol = d)
colnames(betas) <- colnames(X)
# define quantities for the Gibbs sampler
BN <- solve(B0.inv + crossprod(X))
ind0 <- (y == 0) # indicators for zeros
ind1 <- (y == 1) # indicators for ones
# starting values
beta <- rep(0, d)
z <- rep(NA_real_, N)
for (m in seq_len(burnin + M)) {
# Draw z conditional on y and beta
u <- runif(N)
eta <- X %*% beta
pi <- pnorm(eta)
z[ind0] <- eta[ind0] + qnorm(u[ind0] * (1 - pi[ind0]))
z[ind1] <- eta[ind1] + qnorm(1 - u[ind1] * pi[ind1])
# sample beta from the full conditional
bN <- BN %*% (B0inv.b0 + crossprod(X, z))
beta <- t(mvtnorm::rmvnorm(1, mean = bN, sigma = BN))
# Store the beta draws
if (m > burnin) {
betas[m - burnin, ] <- beta
}
}
return(betas)
}We specify the prior on the regression effects as a rather flat Normal independence prior and estimate the model.
set.seed(1234)
betas <- probit(y.unemp, X.unemp, b0 = 0, B0 = 10000)To compute summary statistics from the posterior we use the following function.
res.mcmc <- function(x, lower = 0.025, upper = 0.975) {
y <- c(quantile(x, lower), mean(x), quantile(x, upper))
names(y) <- c(paste0(lower * 100, "%"), "Mean", paste0(upper * 100, "%"))
y
} | intercept | female | age18 | wcollar | unemp97 | |
|---|---|---|---|---|---|
| 2.5% | -2.123 | 0.109 | 0.023 | -0.292 | 2.412 |
| Mean | -1.973 | 0.214 | 0.027 | -0.183 | 2.523 |
| 97.5% | -1.831 | 0.325 | 0.032 | -0.080 | 2.639 |
(p_unemploy_base <- pnorm(res_beta[1, 2]))
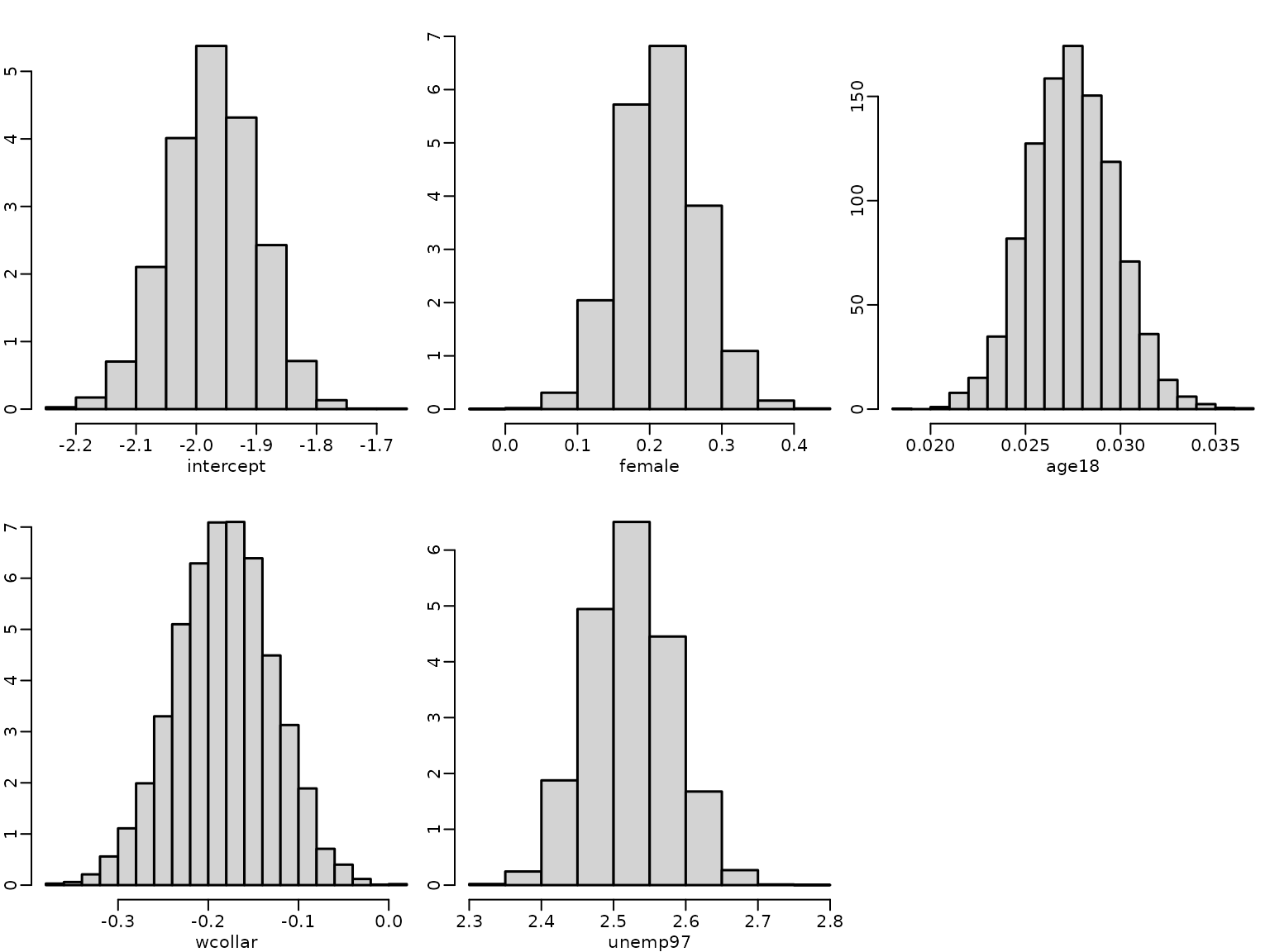
#> [1] 0.5432806The estimated risk of unemployment for a baseline person is low and it is even lower for a white collar worker. It is higher for females, older persons and particularly for those unemployed in 1997.
for (j in seq_len(ncol(betas))) {
hist(betas[, j], freq = FALSE, main = "", xlab = colnames(betas)[j],
ylab = "")
}
A plot of the autocorrelation of the draws shows that although there is some autocorrelation, it vanishes after a few lags.
for (j in seq_len(ncol(betas))) {
acf(betas[, j], main = "", xlab = colnames(betas)[j], ylab = "")
} The sampler is easy to implement, however there might be problems when
the response variable contains either only few or very many successes.
To illustrate this issue, we use data where in
trials only 1 success or only 1 failure is observed.
The sampler is easy to implement, however there might be problems when
the response variable contains either only few or very many successes.
To illustrate this issue, we use data where in
trials only 1 success or only 1 failure is observed.
N <- 500
X <- matrix(1, nrow = N)
y1 <- c(0, rep(1, N-1))
betas1 <- probit(y1, X, b0 = 0, B0 = 10000)
y2 <- c(rep(0, N-1), 1)
betas2 <- probit(y2, X, b0 = 0, B0 = 10000) In both cases the autocorrelation of the draws decreases very slowly.
plot(betas1, type = "l", main = "", xlab = "", ylab = "")
acf(betas1)
plot(betas2, type = "l", main = "", xlab = "", ylab = "")
acf(betas2)
High autocorrelated draws in probit models not only occur if successes or failures are very rare, but also when a covariate (or a linear combination of covariates) perfectly allows to predict successes and/or failures. Complete separation means that both successes and failures can be perfectly predicted by a covariate, whereas with quasi-complete separation only either successes or failures can be predicted perfectly.
To illustrate posterior sampling, in the case of complete separation, we simulate observations with 10 successes and 490 failures. We add a binary predictor where for we observe only successes and for only failures.
N <- 500
ns <- 10
x <- rep(c(1, 0), c(ns, N - ns))
y <- rep(c(1, 0), c(ns, N - ns))
table(y, x)
#> x
#> y 0 1
#> 0 490 0
#> 1 0 10We estimate the model parameters and plot the ACF of the draws. Again the autocorrelations remain high even for lag 35.
X <- cbind(rep(1, N), x)
betas <- probit(y, X, b0 = 0, B0 = 10000)
plot(betas[, 1], type = "l", main = "", xlab = "", ylab = "")
acf(betas[, 1])
plot(betas[, 2], type = "l", main = "", xlab = "", ylab = "")
acf(betas[, 2])
Finally we simulate another data set of observations with 10 successes and 490 failures. In this data for only successes but for successes as well as failures are observed and hence there is quasi-separation.
N <- 500
x <- rep(c(1, 0), c(ns, N-ns))
y <- rep(c(1, 0), each = N/2)
table(y, x)
#> x
#> y 0 1
#> 0 250 0
#> 1 240 10A plot of the acf of the draws shows low autocorrelation for the intercept but autocorrelations for the covariate effect are again high.
X <- cbind(rep(1, N), x)
betas <- probit(y, X, b0 = 0, B0 = 10000)
plot(betas[, 1], type = "l", main = "", xlab = "", ylab = "")
acf(betas[, 1])
plot(betas[, 2], type = "l", main = "", xlab = "", ylab = "")
acf(betas[, 2])
High autocorrelations typically indicate problems with the sampler. If there is complete or quasi-complete separation in the data, the likelihood is monotone and the maximum likelihood estiamte does not exist. In a Bayesian approach using a flat, improper prior on the regression effects will result in an improper posterior distribution. Hence, a proper prior is required to avoid improper posteriors in case of separation.
In the examples above we used a proper prior which is rather flat. With a more informative prior, the autocorrelations of the draws are lower. This can be seen in the next figure, where the simulated data under quasi-separation are re-analyzed with a Normal prior that is tighter around zero.
betas <- probit(y, X, b0 = 0, B0 = 2.5^2)
plot(betas[, 1], type = "l", main = "", xlab = "", ylab = "")
acf(betas[, 1])
plot(betas[, 2], type = "l", main = "", xlab = "", ylab = "")
acf(betas[, 2])
Example 8.4: Labor market data
We now estimate a logistic regression model for the labor market data using the two-block Polya-Gamma sampler.
logit <- function(y, X, b0 = 0, B0 = 10000,
burnin = 1000L, M = 5000L) {
N <- length(y)
d <- ncol(X) # number regression effects
B0.inv <- diag(rep(1 / B0, length.out = d), nrow = d)
b0 <- rep(b0, length.out = d)
B0inv.b0 <- B0.inv %*% b0
betas <- matrix(NA_real_, nrow = M, ncol = d)
colnames(betas) <- colnames(X)
# define quantities for the Gibbs sampler
ind0 <- (y == 0) # indicators for zeros
ind1 <- (y == 1) # indicators for ones
# starting values
beta <- rep(0, d)
z <- rep(NA_real_, N)
omega <-rep(NA_real_, N)
for (m in seq_len(burnin + M)) {
# Draw z conditional on y and beta
eta <- X %*% beta
pi <- plogis(eta)
u <- runif(N)
z[ind0] <- eta[ind0] + qlogis(u[ind0] * (1 - pi[ind0]))
z[ind1] <- eta[ind1] + qlogis (1 - u[ind1] * pi[ind1])
# Draw omega conditional on y, beta and z
omega <- pgdraw::pgdraw(b = 1, c = z - eta)
# sample beta from the full conditional
Xomega <- matrix(omega, ncol = d, nrow = N) * X
BN <- solve(B0.inv + crossprod(Xomega, X))
bN <- BN %*% (B0inv.b0 + crossprod(Xomega, z))
beta <- t(mvtnorm::rmvnorm(1, mean = bN, sigma = BN))
# Store the beta draws
if (m > burnin) {
betas[m - burnin, ] <- beta
}
}
return(betas)
}We again use the flat independence Normal prior on the regression effects and estimate the model.
betas_logit <- logit(y.unemp, X.unemp, b0 = 0, B0 = 10000)
print(str(betas_logit))
#> num [1:5000, 1:5] -3.32 -3.43 -3.57 -3.53 -3.68 ...
#> - attr(*, "dimnames")=List of 2
#> ..$ : NULL
#> ..$ : chr [1:5] "intercept" "female" "age18" "wcollar" ...
#> NULL
res_beta_logit <- apply(betas_logit, 2, res.mcmc)
knitr::kable(round(res_beta_logit, 3))| intercept | female | age18 | wcollar | unemp97 | |
|---|---|---|---|---|---|
| 2.5% | -4.072 | 0.133 | 0.044 | -0.611 | 4.091 |
| Mean | -3.659 | 0.399 | 0.056 | -0.344 | 4.370 |
| 97.5% | -3.281 | 0.658 | 0.067 | -0.067 | 4.669 |
(p_unemploy_base <- plogis(res_beta_logit[1, 2]))
#> [1] 0.5332866Note that the logistic distribution has a variance of and hence the regression effects are absolutely larger than in the probit model. However any probability computed from the two models will be very close, e.g., compare the probability to be unemployed for the baseline person.
We can compare the posterior estimates of the coefficients in the probit model those of of logit model by multiplying them with and see that there is not much difference.
| intercept | female | age18 | wcollar | unemp97 | |
|---|---|---|---|---|---|
| 2.5% | -3.851 | 0.197 | 0.042 | -0.529 | 4.376 |
| Mean | -3.578 | 0.388 | 0.050 | -0.331 | 4.575 |
| 97.5% | -3.321 | 0.589 | 0.058 | -0.145 | 4.787 |
Section 8.2: Count response variables
Example 8.5: Road safety data
We fit two different Poisson regression models:
a small model with intercept, intervention effect and holiday dummy (activated in July/August);
a large model with intercept, intervention effect, linear trend, and a seasonal pattern captured by monthly dummies.
The sampler performance for these two models is assessed to study how the acceptance rate deteroriates, if increases.
We load the data and extract the observations for the children in Linz.
data("accidents", package = "BayesianLearningCode")
y <- accidents[, "children_accidents"]
e <- rep(1, length(y))Then, we define the regressor matrix.
X <- cbind(intercept = rep(1, length(y)),
intervention = rep(c(0, 1), c(7 * 12 + 9, 8 * 12 + 3)),
holiday = rep(rep(c(0, 1, 0), c(6, 2, 4)), 16))To compute the parameters of the Normal proposal density, we use the Newton-Raphson estimator described in Section 8.2.1.
gen.proposal.poisson <- function(y, X, e, b0 = 0, B0 = 100, t.max = 30) {
N <- length(y)
d <- ncol(X)
betas <- matrix(NA_real_, ncol = t.max, nrow = d)
beta.old <- c(log(mean(y)), rep(0, d - 1))
b0 <- rep(b0, length.out = d)
B0.inv <- diag(rep(1 / B0, length.out = d), nrow = d)
for (t in 1:t.max) {
rate <- e * exp(X %*% beta.old)
score <- t(crossprod(y - rate, X) - (beta.old - b0) %*% B0.inv)
H <- -B0.inv
for (i in 1:N) {
H <- H - rate[i] * tcrossprod(X[i, ])
}
betas[, t] <- beta.old - solve(H, score)
}
qmean <- betas[, t.max]
# determine the variance matrix
rate <- e * exp(X %*% qmean)
H <- -B0.inv
for (i in 1:N) {
H <- H - rate[i] * tcrossprod(X[i, ])
}
qvar <- -solve(H)
return(parms.proposal = list(mean = qmean,
var = qvar))
}We use a flat Normal prior on the regression effects.
parms.proposal <- gen.proposal.poisson(y, X, e, b0 = 0, B0 = 100)
print(parms.proposal)
#> $mean
#> [1] 0.8867173 -0.3465599 -0.5944486
#>
#> $var
#> [,1] [,2] [,3]
#> [1,] 0.005121126 -0.0048204767 -0.0031235498
#> [2,] -0.004820477 0.0111451181 0.0002039249
#> [3,] -0.003123550 0.0002039249 0.0303637113Next we set up the independence Metropolis-Hastings algorithm and estimate the model parameters.
poisson <- function(y, X, e, b0 = 0, B0 = 100, qmean, qvar,
burnin = 1000L, M = 5000L) {
d <- ncol(X)
beta.post <- matrix(ncol = d, nrow = M)
colnames(beta.post) <- colnames(X)
acc <- numeric(length = M)
b0 <- rep(b0, length.out = d)
B0 <- diag(rep(B0, length.out = d), nrow = d)
beta <- as.vector(mvtnorm::rmvnorm(1, mean = qmean, sigma = qvar))
for (m in seq_len(burnin + M)) {
beta.old <- beta
beta.proposed <- as.vector(mvtnorm::rmvnorm(1, mean = qmean, sigma = qvar))
# compute log proposal density at proposed and old value
lq_proposed <- mvtnorm::dmvnorm(beta.proposed, mean = qmean, sigma = qvar,
log = TRUE)
lq_old <- mvtnorm::dmvnorm(beta.old, mean = qmean, sigma = qvar,
log = TRUE)
# compute log prior of proposed and old value
lpri_proposed <- mvtnorm::dmvnorm(beta.proposed, mean = b0, sigma = B0,
log = TRUE)
lpri_old <- mvtnorm::dmvnorm(beta.old, mean = b0, sigma = B0,
log = TRUE)
# compute loglikelihood of proposed and old value
lh_proposed <- dpois(y, e * exp(X %*% beta.proposed), log = TRUE)
lh_old <- dpois(y, e * exp(X %*% beta.old), log = TRUE)
maxlik <- max(lh_old, lh_proposed)
ll <- sum(lh_proposed - maxlik) - sum(lh_old - maxlik)
# compute acceptance probability and accept or not
log_acc <- min(0, ll + lpri_proposed - lpri_old + lq_old - lq_proposed)
if (log(runif(1)) < log_acc) {
beta <- beta.proposed
accept <- 1
} else {
beta <- beta.old
accept <- 0
}
# Store the beta draws
if (m > burnin) {
beta.post[m-burnin, ] <- beta
acc[m-burnin] <- accept
}
}
return(res = list(beta.post = beta.post, accept = mean(acc)))
}
res1 <- poisson(y, X, e, b0 = 0, B0 = 100,
qmean = parms.proposal$mean, qvar = parms.proposal$var)
res.poisson1 <- t(rbind(apply(res1$beta.post, 2, res.mcmc),
`exp(Mean)` = exp(colMeans(res1$beta.post))))
knitr::kable(round(res.poisson1, 3))| 2.5% | Mean | 97.5% | exp(Mean) | |
|---|---|---|---|---|
| intercept | 0.741 | 0.864 | 0.996 | 2.373 |
| intervention | -0.526 | -0.343 | -0.142 | 0.710 |
| holiday | -1.172 | -0.815 | -0.431 | 0.443 |
print(res1$accept)
#> [1] 0.3468We then fit an alternative model with intercept, intervention effect, linear trend and saisonal dummy variables.
seas <- rbind(diag(1, 11), rep(-1, 11))
seas.dummies <- matrix(rep(t(seas), 16), ncol = 11, byrow = TRUE)
colnames(seas.dummies) <- c("Jan", "Feb", "Mar", "Apr","May", "Jun", "Jul",
"Aug", "Sep", "Oct", "Nov")
X.large <- cbind(X,
lin.trend = 1:length(y),
seas.dummies)We set the prior parameters and compute parameters of the proposal distribution
parms.proposal2 <- gen.proposal.poisson(y, X.large, e, b0 = 0, B0 = 100)and fit the model.
res2 <- poisson(y, X.large, e, b0 = 0, B0 = 100,
qmean = parms.proposal2$mean, qvar = parms.proposal2$var)
res.poisson2 <- t(rbind(apply(res2$beta.post, 2, res.mcmc),
`exp(Mean)` = exp(colMeans(res2$beta.post))))
knitr::kable(round(res.poisson2, 3))| 2.5% | Mean | 97.5% | exp(Mean) | |
|---|---|---|---|---|
| intercept | -1.261 | 0.787 | 2.993 | 2.196 |
| intervention | -0.727 | -0.337 | 0.007 | 0.714 |
| holiday | -12.898 | -0.424 | 11.692 | 0.654 |
| lin.trend | -0.003 | 0.000 | 0.004 | 1.000 |
| Jan | -1.940 | 0.208 | 2.179 | 1.232 |
| Feb | -2.504 | -0.432 | 1.659 | 0.649 |
| Mar | -2.153 | 0.022 | 2.213 | 1.022 |
| Apr | -1.932 | 0.291 | 2.325 | 1.337 |
| May | -2.666 | -0.315 | 1.955 | 0.730 |
| Jun | -1.895 | 0.326 | 2.428 | 1.385 |
| Jul | -10.521 | -0.203 | 10.299 | 0.816 |
| Aug | -10.608 | -0.330 | 10.104 | 0.719 |
| Sep | -2.248 | -0.052 | 2.026 | 0.950 |
| Oct | -1.988 | 0.362 | 2.439 | 1.436 |
| Nov | -2.357 | -0.007 | 2.100 | 0.993 |
print(res2$accept)
#> [1] 0.1934
# xtable(res.poisson1, caption= "Count Data: estimation results for Model 1", label="res.count1", digits=3)
# xtable(res.poisson2, caption= "Count Data: estimation results for Model 2", label="res.count2", digits=3)Example 8.6: Road safety data
Now we analyse the road safety data allowing for unobserved heterogeneity. We first set up both the two versions of the three-block MH-within-Gibbs sampler
negbin <- function(y, X, e, b0 = 0, B0 = 100, qmean, qvar, pri.alpha,
full.gibbs= FALSE, burnin = 1000L, M = 50000L) {
N=length(y)
d <- ncol(X)
beta.post <- matrix(ncol = d, nrow = M)
colnames(beta.post) <- colnames(X)
b0 <- rep(b0, length.out = d)
B0 <- diag(rep(B0, length.out = d), nrow = d)
acc.beta <- numeric(length = M)
alpha.post<- rep(NA,M)
acc.alpha <-rep(NA,M)
c_alpha <- 0.1
# set starting values
beta <- as.vector(mvtnorm::rmvnorm(1, mean = qmean, sigma = qvar))
alpha <- pri.alpha$shape/pri.alpha$rate
phi <- rep(1.,N)
for (m in seq_len(burnin + M)){
# Step 1: draw beta
beta.old <- beta
beta.proposed <- as.vector(mvtnorm::rmvnorm(1, mean = qmean, sigma = qvar))
# compute log proposal density at proposed and old value
lq_proposed <- mvtnorm::dmvnorm(beta.proposed, mean = qmean, sigma = qvar,
log = TRUE)
lq_old <- mvtnorm::dmvnorm(beta.old, mean = qmean, sigma = qvar,
log = TRUE)
# compute log prior of proposed and old value
lpri_proposed <- mvtnorm::dmvnorm(beta.proposed, mean = b0, sigma = B0,
log = TRUE)
lpri_old <- mvtnorm::dmvnorm(beta.old, mean = b0, sigma = B0, log = TRUE)
# compute log likelihood of proposed and old value
lh_proposed <- dpois(y, exp(X %*% beta.proposed), log = TRUE)
lh_old <- dpois(y, exp(X %*% beta.old), log = TRUE)
maxlik <- max(lh_old,lh_proposed)
ll <- sum(lh_proposed - maxlik) - sum(lh_old - maxlik)
# compute acceptance probability and accept or not
log_acc <- min(0, ll + lpri_proposed - lpri_old + lq_old - lq_proposed)
if (log(runif(1)) < log_acc) {
beta <- beta.proposed
acc.b <- 1
}else{
beta <- beta.old
acc.b <- 0
}
linpred <- X %*% beta
# Step 2: Sample alpha
alpha.old <- alpha
alpha.proposed <- alpha.old * exp(c_alpha * rnorm(1))
if (full.gibbs) {
llik_alpha.proposed <- sum(dgamma(phi, shape = alpha.proposed,
rate = alpha.proposed, log = TRUE))
llik_alpha.old <- sum(dgamma(phi, shape = alpha.old,
rate = alpha.old, log = TRUE))
} else {
llik_alpha.proposed <- sum(dnbinom(y, size = alpha.proposed,
mu = e * exp(linpred), log = TRUE))
llik_alpha.old <- sum(dnbinom(y, size = alpha.old,
mu = e * exp(linpred), log = TRUE))
}
log_acc_alpha <- llik_alpha.proposed - llik_alpha.old +
dgamma(alpha.proposed, shape = pri.alpha$shape,
rate = pri.alpha$rate, log = TRUE) -
dgamma(alpha.old, shape = pri.alpha$shape,
rate = pri.alpha$rate,log=TRUE) +
log(alpha.proposed) - log(alpha.old)
if (log(runif(1)) < log_acc_alpha) {
alpha <- alpha.proposed
acc.a <- 1
} else {
alpha <- alpha.old
acc.a <- 0
}
# Step 3: sample phi from its full conditional
phi <- rgamma(N, shape = alpha + y, rate = alpha + e * exp(linpred))
# Save the draws
if (m > burnin) {
beta.post[m - burnin, ] <- beta
acc.beta[m - burnin] <- acc.b
alpha.post[m - burnin] <- alpha
acc.alpha[m - burnin] <- acc.a
}
}
return(res = list(beta.post = beta.post, acc.beta = acc.beta,
alpha.post = alpha.post,acc.alpha = acc.alpha))
}We specify the prior on as the Gamma distribution with shape 2 and rate 0.5 and use both samplers to estimate the model parameters.
pri.alpha <- data.frame(shape = 2, rate = 0.5)
res1 <- negbin(y, X, e, qmean = parms.proposal$mean, qvar = parms.proposal$var,
pri.alpha = pri.alpha, full.gibbs = TRUE)
res.negbin.full <- cbind(apply(res1$beta.post, 2, res.mcmc),
res.mcmc(res1$alpha.post))
colnames(res.negbin.full)[4] <- "alpha"
knitr::kable(round(res.negbin.full, 3))| intercept | intervention | holiday | alpha | |
|---|---|---|---|---|
| 2.5% | 0.721 | -0.567 | -1.204 | 6.512 |
| Mean | 0.865 | -0.348 | -0.792 | 12.370 |
| 97.5% | 1.005 | -0.143 | -0.424 | 20.922 |
res2 <- negbin(y, X, e, qmean = parms.proposal$mean, qvar = parms.proposal$var,
pri.alpha = pri.alpha, full.gibbs = FALSE)
res.negbin.partial <- cbind(apply(res2$beta.post, 2, res.mcmc),
res.mcmc(res2$alpha.post))
colnames(res.negbin.partial)[4] <- "alpha"
knitr::kable(round(res.negbin.partial, 3))| intercept | intervention | holiday | alpha | |
|---|---|---|---|---|
| 2.5% | 0.723 | -0.557 | -1.164 | 6.602 |
| Mean | 0.866 | -0.350 | -0.785 | 12.439 |
| 97.5% | 1.004 | -0.135 | -0.431 | 20.799 |
As expected estimation results using both samplers are rather similar.
#require(xtable)
#xtable(cbind(t(res.negbin.full), t(res.negbin.partial)), label="exam:negbin_est")Section 8.2.3: Evaluating MCMC samplers
print(c(mean(res1$acc.beta), mean(res1$acc.alpha)))
#> [1] 0.35236 0.70344
print(c(mean(res2$acc.beta), mean(res2$acc.alpha)))
#> [1] 0.35816 0.89920
if (pdfplots) {
pdf("8-2_1.pdf", width = 8, height = 5)
}
par(mfrow = c(1,2), mar = c(2.5, 2.5, 1.5, .1), mgp = c(1.5, .5, 0), lwd = 1.5)
qqplot(res1$beta.post[,1], res2$beta.post[,1], xlab = "Full Gibbs",
ylab = "Partial Gibbs", main = "Intercept")
abline(a = 0, b = 1)
qqplot(res1$alpha.post, res2$alpha.post, xlab = "Full Gibbs",
ylab = "Partial Gibbs", main = "Alpha")
abline(a = 0, b = 1)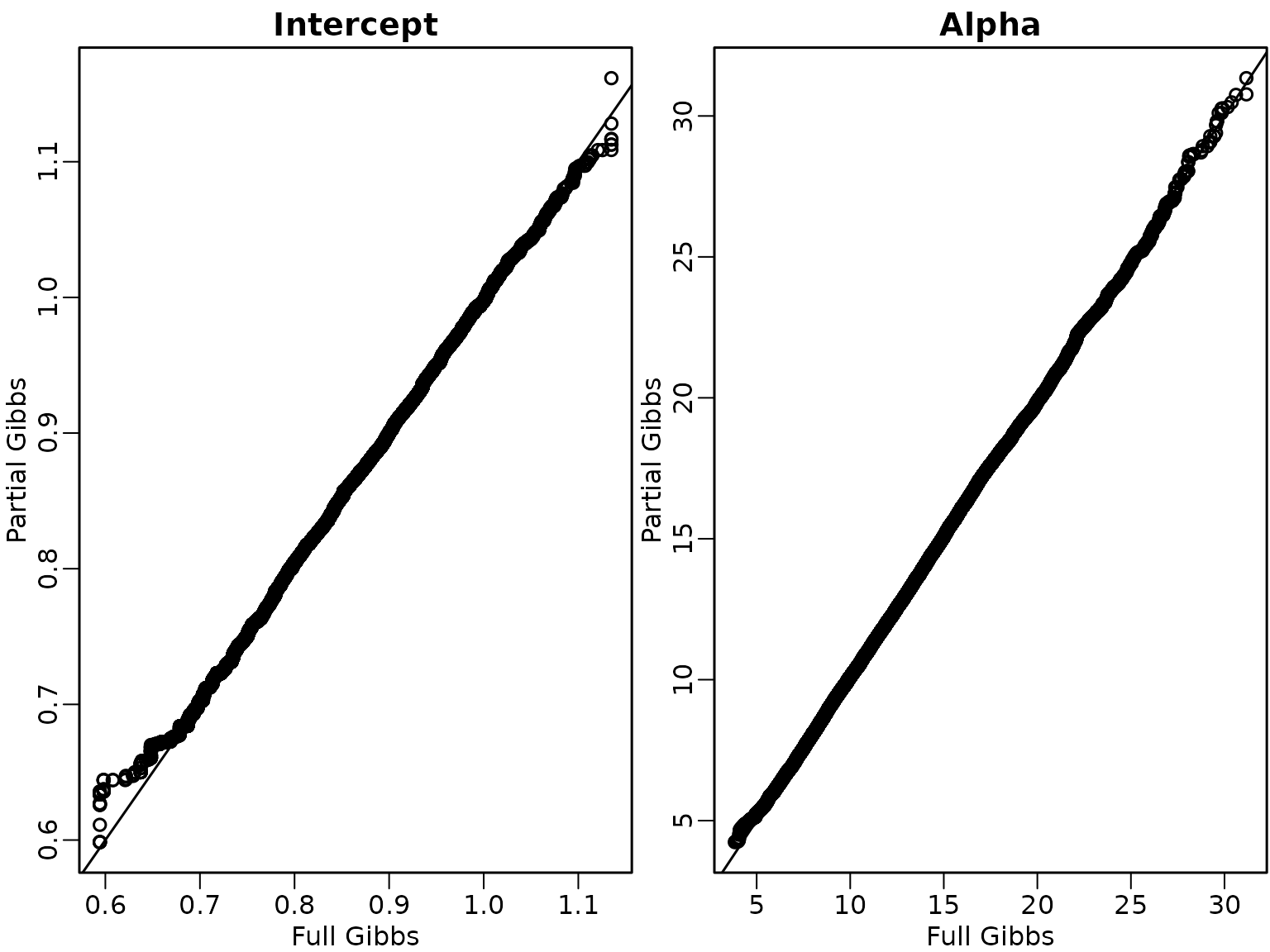
Section 8.3: Beyond i.i.d. Gaussian error distributions
Example 8.12: Star cluster data
The bivariate data set of the star cluster CYG OB1 is available in package robustbase and we load it from this package and visualize it in a scatter plot:
data("starsCYG", package = "robustbase")
plot(starsCYG, pch = 19, xlim = c(3, 5), ylim = c(3, 7),
xlab = "log temperature", ylab = "log light intensity")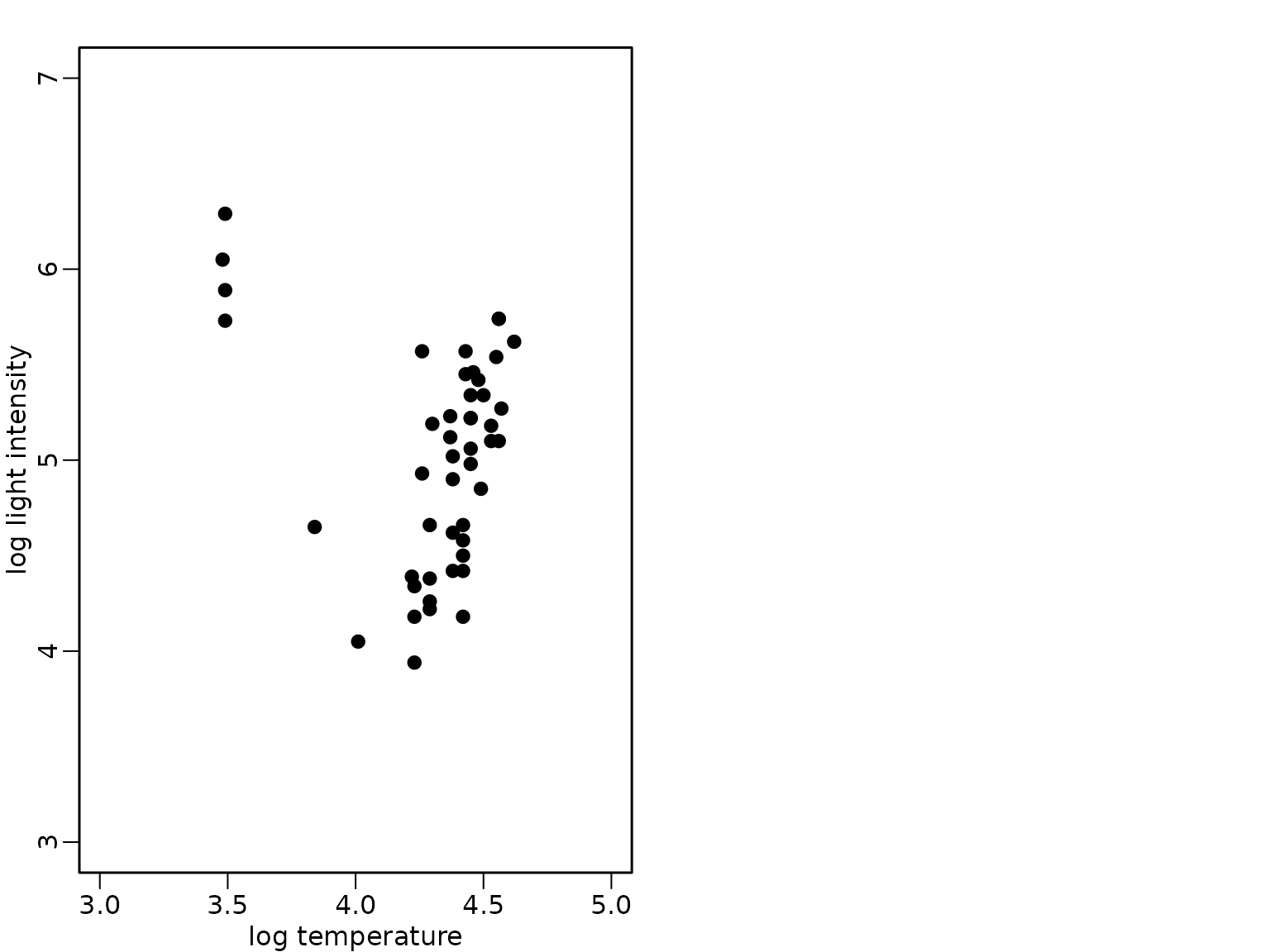
The four giant stars which can also be identified in the scatter plot have the following indices in the data set:
index <- c(11, 20, 30, 34)We fit a standard Bayesian regression analysis under the improper prior and determine the mean and pointwise 95%-HPD regions of the posterior predictive distribution using (a) the full data set and (b) the data set where the observations , , , corresponding to the giant stars are omitted.
The posterior predictive distribution for a single observation with covariate value given the sample with model matrix is available in closed form when using the improper prior and corresponds to the prediction intervals obtained using OLS estimation.
ols_all <- lm(log.light ~ log.Te, data = starsCYG)
xnew <- seq(3, 5, length.out = 100)
preds_all <- predict(ols_all, newdata = data.frame(log.Te = xnew),
interval = "prediction")
ols_subset <- lm(log.light ~ log.Te, data = starsCYG[-index, ])
preds_subset <- predict(ols_subset, newdata = data.frame(log.Te = xnew),
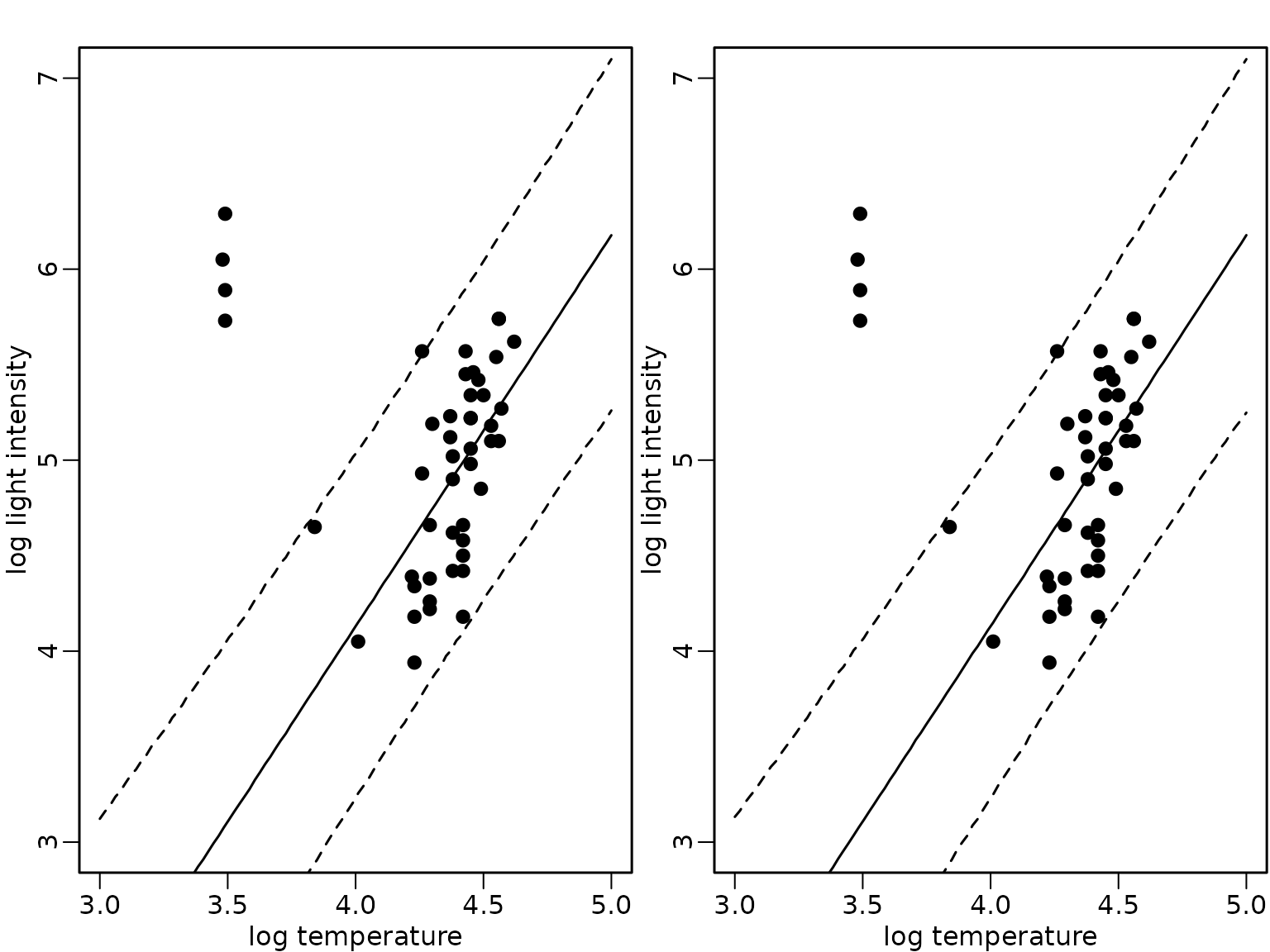
interval = "prediction")We compare the expected values (full lines) and the pointwise 95%-HPD regions in the following fiugre for the model fit using all data (left) and only the subset without the giant stars (right).
plot(starsCYG, pch = 19, xlim = c(3, 5), ylim = c(3, 7),
xlab = "log temperature", ylab = "log light intensity")
lines(xnew, preds_all[, "fit"])
lines(xnew, preds_all[, "lwr"], lty = 2)
lines(xnew, preds_all[, "upr"], lty = 2)
plot(starsCYG, pch = 19, xlim = c(3, 5), ylim = c(3, 7),
xlab = "log temperature", ylab = "log light intensity")
lines(xnew, preds_subset[, "fit"])
lines(xnew, preds_subset[, "lwr"], lty = 2)
lines(xnew, preds_subset[, "upr"], lty = 2)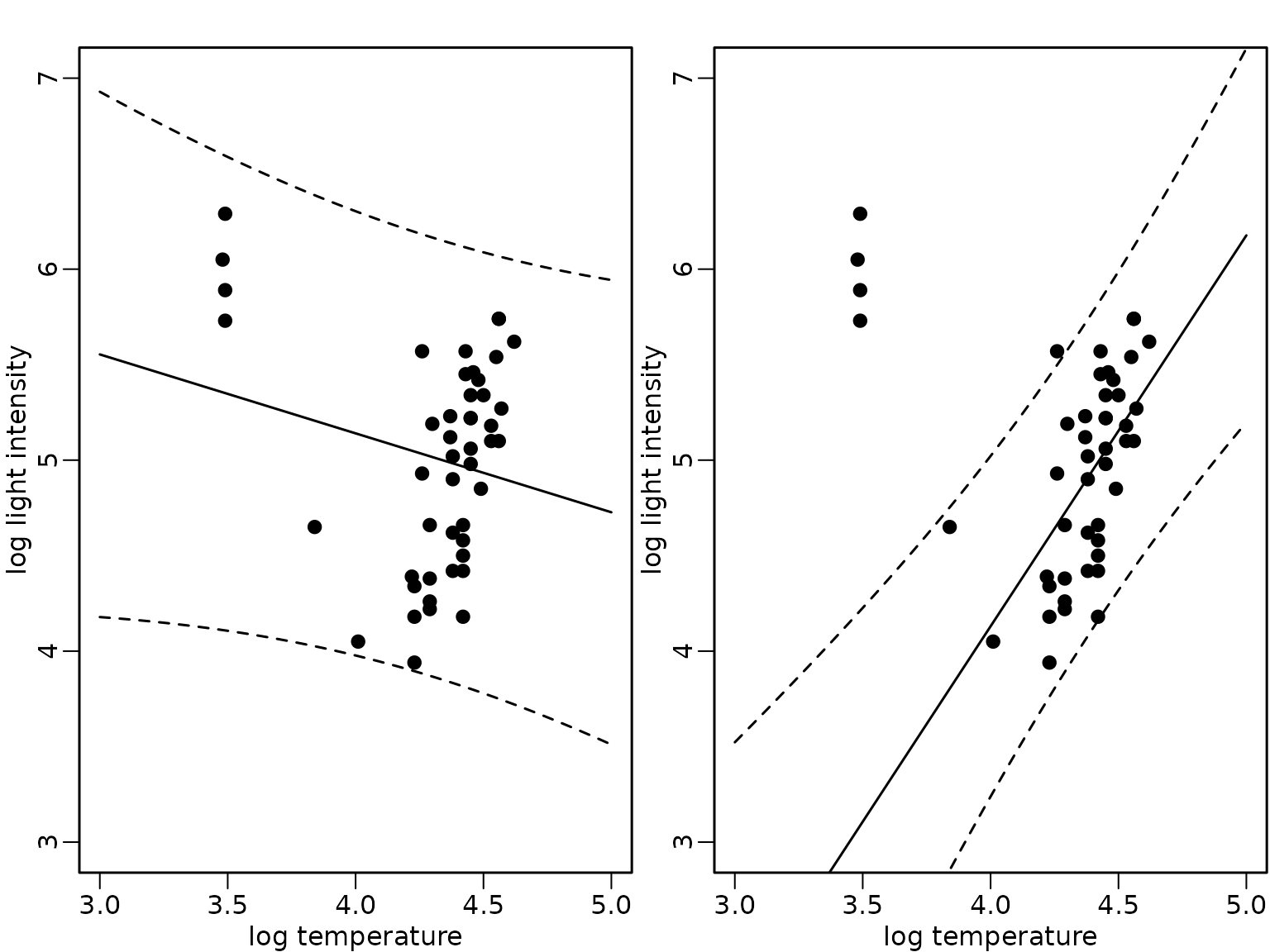
Example 8.13: Star cluster data - heteroscedastic regression with fixed
We define the binary indicator indicating outlying observations, i.e., in this case observations corresponding to giant stars.
We prepare the model matrix and the vector of the response and define the dimensions.
For the heteroscedastic regression, we define weights depending on the binary indicator which are either 1 or equal to .
phi <- 0.001
w <- phi^(S - 1)We include the weights and update the Gibbs sampling scheme for the linear regression defined in Chapter 6 accordingly.
set.seed(1)
# define prior parameters of semi-conjugate prior
B0inv <- diag(rep(1 / 10000, d), nrow = d)
b0 <- rep(0, d)
c0 <- 2.5
C0 <- 1.5
# define quantities for the Gibbs sampler taking the weights into
# account
Xtilde <- sqrt(w) * X
ytilde <- sqrt(w) * y
wXX <- crossprod(Xtilde)
wXy <- t(Xtilde) %*% ytilde
cN <- c0 + N / 2
# define burnin and M
burnin <- 1000
M <- 100000
# prepare storing of results
betas <- matrix(NA_real_, nrow = burnin + M, ncol = d)
sigma2s <- rep(NA_real_, burnin + M)
colnames(betas) <- colnames(X)
# starting value for sigma2
sigma2 <- var(y) / 2
for (m in 1:(burnin + M)) {
# sample beta from the full conditional
BN <- solve(B0inv + wXX / sigma2)
bN <- BN %*% (B0inv %*% b0 + wXy / sigma2)
beta <- t(mvtnorm::rmvnorm(1, mean = bN, sigma = BN))
# sample sigma^2 from its full conditional
eps <- ytilde - Xtilde %*% beta
CN <- C0 + crossprod(eps) / 2
sigma2 <- rinvgamma(1, cN, CN)
betas[m, ] <- beta
sigma2s[m] <- sigma2
}Based on the posterior draws of the parameters we determine draws
from the predictive distributions for new observations with
xnew values:
pred_hetero <- sapply(1:M, function(m) {
rnorm(length(xnew), cbind(1, xnew) %*% betas[m, ], sqrt(sigma2s[m]))
})We visualize again the mean and the 95%-HPD region together with the data points and show that the fit now is robust to the outlying observations.
plot(starsCYG, pch = 19, xlim = c(3, 5), ylim = c(3, 7),
xlab = "log temperature", ylab = "log light intensity")
lines(xnew, rowMeans(pred_hetero))
lines(xnew, apply(pred_hetero, 1, quantile, 0.025), lty = 2)
lines(xnew, apply(pred_hetero, 1, quantile, 0.975), lty = 2)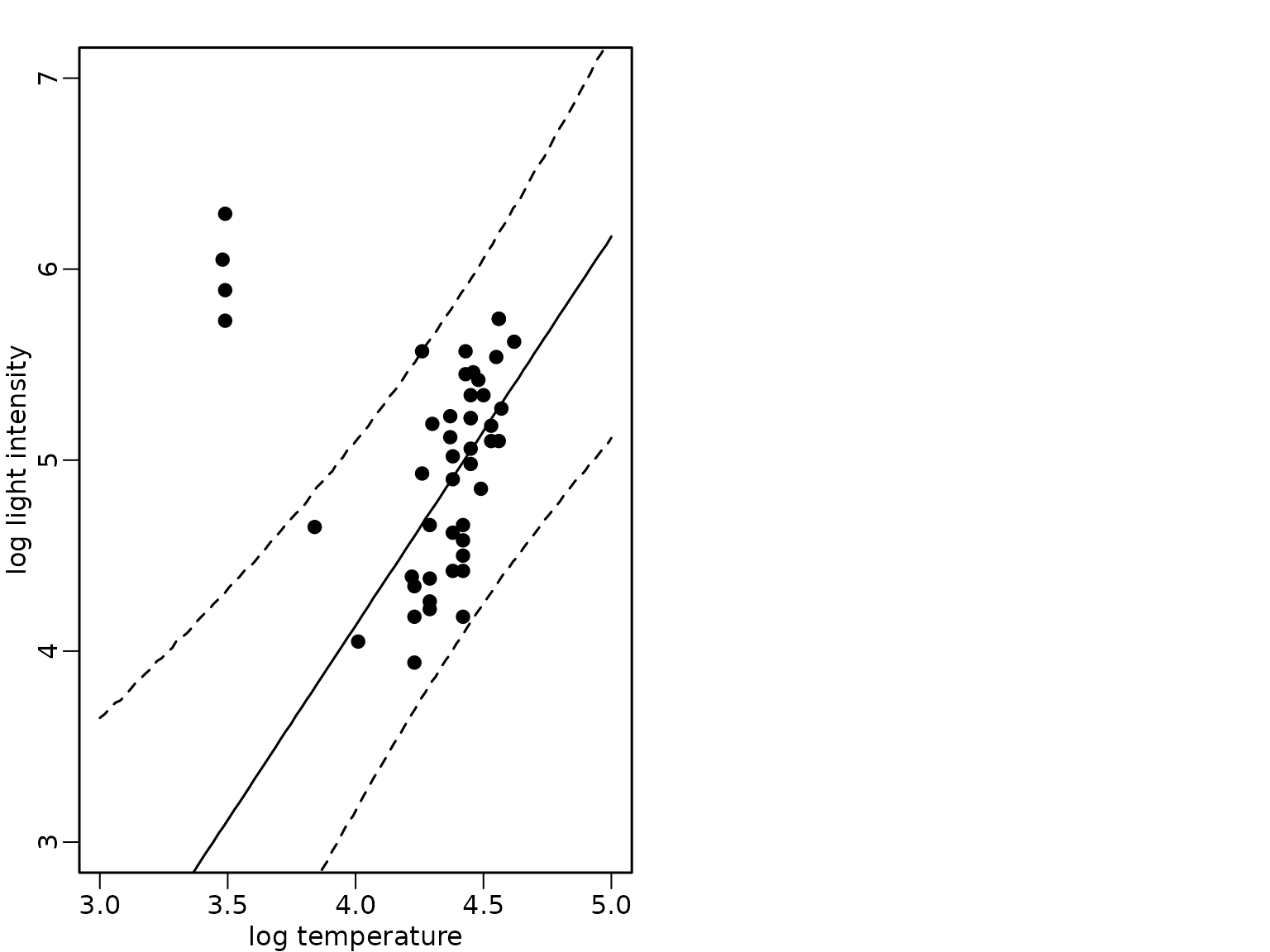
Example 8.14: Star cluster data - two-component Gaussian mixture with fixed and
We now assume that the indices of the giant stars are not known. We only assume that a two-component mixture is used as weight distribution where the weights are given by and the indicators are unknown. But we assume that the proportion of giant stars is known and we assume this to correspond to 10%.
eta <- 0.1
phi <- 0.001
# define prior parameters of semi-conjugate prior
B0inv <- diag(rep(1, d), nrow = d)
b0 <- coef(ols_subset)We now modify the Gibbs sampling code to include a sampling step for the mixture component indicators.
set.seed(1)
# starting values for beta and sigma2
beta <- coef(ols_subset)
sigma2 <- var(y) / 2
for (m in seq_len(burnin + M)) {
# mixture component indicator
Xbeta <- X %*% beta
posterior <- cbind((1 - eta) * dnorm(y, Xbeta, sqrt(sigma2)),
eta * dnorm(y, Xbeta, sqrt(sigma2 / phi)))
posterior <- posterior / rowSums(posterior)
S <- 1 + rbinom(nrow(posterior), prob = posterior[, 2],
size = 1)
# re-weight
w <- phi^(S - 1)
Xtilde <- sqrt(w) * X
ytilde <- sqrt(w) * y
wXX <- crossprod(Xtilde)
wXy <- t(Xtilde) %*% ytilde
# sample beta from the full conditional
BN <- solve(B0inv + wXX / sigma2)
bN <- BN %*% (B0inv %*% b0 + wXy / sigma2)
beta <- t(mvtnorm::rmvnorm(1, mean = bN, sigma = BN))
# sample sigma^2 from its full conditional
eps <- ytilde - Xtilde %*% beta
CN <- C0 + crossprod(eps) / 2
sigma2 <- rinvgamma(1, cN, CN)
betas[m, ] <- beta
sigma2s[m] <- sigma2
}
preds_mix_1 <- sapply(1:M, function(m) {
rnorm(length(xnew), cbind(1, xnew) %*% betas[m, ], sqrt(sigma2s[m]))
})We visualize again the mean and the 95%-HPD region together with the data points and show that the fit now is robust to the outlying observations.
Example 8.14: Star cluster data - two-component Gaussian mixture with fixed
We now assume that the indices of the giant stars are not known. We only assume that a two-component mixture is used as weight distribution where the weights are given by and the indicators are unkown. Now, we also assume that the proportion of giant stars is unknown.
We need to specify a prior for . The usual prior is a beta prior and we will use a prior which has mean 0.1 and corresponds to a prior sample size of 10.
a0 <- 1
d0 <- 9We now modify the Gibbs sampling code to also include a sampling step for the component size.
set.seed(1)
# prepare storing of results
etas <- rep(NA_real_, burnin + M)
# starting values for eta, beta and sigma2
eta <- 0.1
beta <- coef(ols_subset)
sigma2 <- var(y) / 2
for (m in seq_len(burnin + M)) {
# mixture component indicator
Xbeta <- X %*% beta
posterior <- cbind((1 - eta) * dnorm(y, Xbeta, sqrt(sigma2)),
eta * dnorm(y, Xbeta, sqrt(sigma2 / phi)))
posterior <- posterior / rowSums(posterior)
S <- 1 + rbinom(nrow(posterior), prob = posterior[, 2],
size = 1)
# sample eta
aN <- a0 + sum(S == 2)
dN <- d0 + sum(S == 1)
eta <- rbeta(1, aN, dN)
# re-weight
w <- phi^(S - 1)
Xtilde <- sqrt(w) * X
ytilde <- sqrt(w) * y
wXX <- crossprod(Xtilde)
wXy <- t(Xtilde) %*% ytilde
# sample beta from the full conditional
BN <- solve(B0inv + wXX / sigma2)
bN <- BN %*% (B0inv %*% b0 + wXy / sigma2)
beta <- t(mvtnorm::rmvnorm(1, mean = bN, sigma = BN))
# sample sigma^2 from its full conditional
eps <- ytilde - Xtilde %*% beta
CN <- C0 + crossprod(eps) / 2
sigma2 <- rinvgamma(1, cN, CN)
betas[m, ] <- beta
sigma2s[m] <- sigma2
etas[m] <- eta
}
preds_mix_2 <- sapply(1:M, function(m) {
rnorm(length(xnew), cbind(1, xnew) %*% betas[m, ], sqrt(sigma2s[m]))
})
plot(starsCYG, pch = 19, xlim = c(3, 5), ylim = c(3, 7),
xlab = "log temperature", ylab = "log light intensity")
lines(xnew, rowMeans(preds_mix_1))
lines(xnew, apply(preds_mix_1, 1, quantile, 0.025), lty = 2)
lines(xnew, apply(preds_mix_1, 1, quantile, 0.975), lty = 2)
plot(starsCYG, pch = 19, xlim = c(3, 5), ylim = c(3, 7),
xlab = "log temperature", ylab = "log light intensity")
lines(xnew, rowMeans(preds_mix_2))
lines(xnew, apply(preds_mix_2, 1, quantile, 0.025), lty = 2)
lines(xnew, apply(preds_mix_2, 1, quantile, 0.975), lty = 2)