Chapter 1: From Bayes’ Rule to Bayes’ Theorem
Chapter01.RmdSection 1.3: Naive Bayes classifiers
Example 1.3 and 1.4: Classifying categorical observations into two states
We have two firms, a reliable firm with and a less reliable firm with .
PrA <- 0.7
PrAC <- 1 - PrAThe failure rate, i.e., the probability of a faulty item, is 0.01 for the reliable firm and 0.05 for the less reliable firm .
PrFA <- 0.01
PrFAC <- 0.05Assume that we observe any number of failures between 0 and 6 (from observations in total), but we don’t know whether the items are from company or .
N <- 100
y <- 0:6We can now compute the probability of dealing with items from company by using Bayes’ theorem.
theta1_y_unnormalized <- PrFA ^ y * (1 - PrFA) ^ (N - y) * PrA
theta0_y_unnormalized <- PrFAC ^ y * (1 - PrFAC) ^ (N - y) * PrAC
normalizer <- theta1_y_unnormalized + theta0_y_unnormalized
res <- rbind("reliable company" = theta1_y_unnormalized / normalizer,
"less reliable company" = theta0_y_unnormalized / normalizer)
colnames(res) <- y
knitr::kable(round(res, 3))| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| reliable company | 0.993 | 0.965 | 0.842 | 0.505 | 0.164 | 0.036 | 0.007 |
| less reliable company | 0.007 | 0.035 | 0.158 | 0.495 | 0.836 | 0.964 | 0.993 |
Note that, of course, the probabilities of dealing with items from company and are complementary (i.e., sum to one).
Assuming we do not have any prior information about the probability of occurrence of company , we could simply set and recompute the posterior probabilities.
PrA <- 0.5
PrAC <- 1 - PrA
theta1_y_unnormalized <- PrFA ^ y * (1 - PrFA) ^ (N - y) * PrA
theta0_y_unnormalized <- PrFAC ^ y * (1 - PrFAC) ^ (N - y) * PrAC
normalizer <- theta1_y_unnormalized + theta0_y_unnormalized
res <- rbind(theta1_y_unnormalized/ normalizer,
theta0_y_unnormalized/ normalizer)
colnames(res) <- y
rownames(res) <- c("reliable company", "less reliable company")
knitr::kable(round(res, 3))| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| reliable company | 0.984 | 0.922 | 0.695 | 0.304 | 0.077 | 0.016 | 0.003 |
| less reliable company | 0.016 | 0.078 | 0.305 | 0.696 | 0.923 | 0.984 | 0.997 |
Example 1.5: Classifying continuous observations into two states
We have a Bernoulli random variable with prior probability and we know for the females that and for the males that where , , and .
Prtheta = 0.55
lambda = 1/2.2
alpha = 5
beta = 5/5.5We now plot the conditional densities for . Then, we compute the non-normalized posteriors for , mark those in the plot, and add lines to visualize the non-normalized posteriors.
y_all <- seq(0, 10, 0.2)
plot(y_all, dexp(y_all, lambda), type = "l", xlab = "Distance", ylab = "",
main = "Conditional densities and non-normalized posteriors")
lines(y_all, dgamma(y_all, alpha, beta), col = "red")
y <- c(1, 2, 3, 6, 8)
theta1_y_unnormalized <- dexp(y, lambda) * Prtheta
theta0_y_unnormalized <- dgamma(y, alpha, beta) * (1 - Prtheta)
points(y, theta1_y_unnormalized)
points(y, theta0_y_unnormalized, col = "red", pch = 2)
theta1_y_all_unnormalized <- dexp(y_all, lambda) * Prtheta
theta0_y_all_unnormalized <- dgamma(y_all, alpha, beta) * (1 - Prtheta)
lines(y_all, theta1_y_all_unnormalized, lty = 2)
lines(y_all, theta0_y_all_unnormalized, lty = 2, col = "red")
legend("topright", c("Conditional density (female)",
"Conditional density (male)",
"Non-normalized posterior (female)",
"Non-normalized posterior (male)"),
col = rep(c("black", "red"), 2), lty = rep(1:2, each = 2),
pch = c(NA, NA, 1, 2))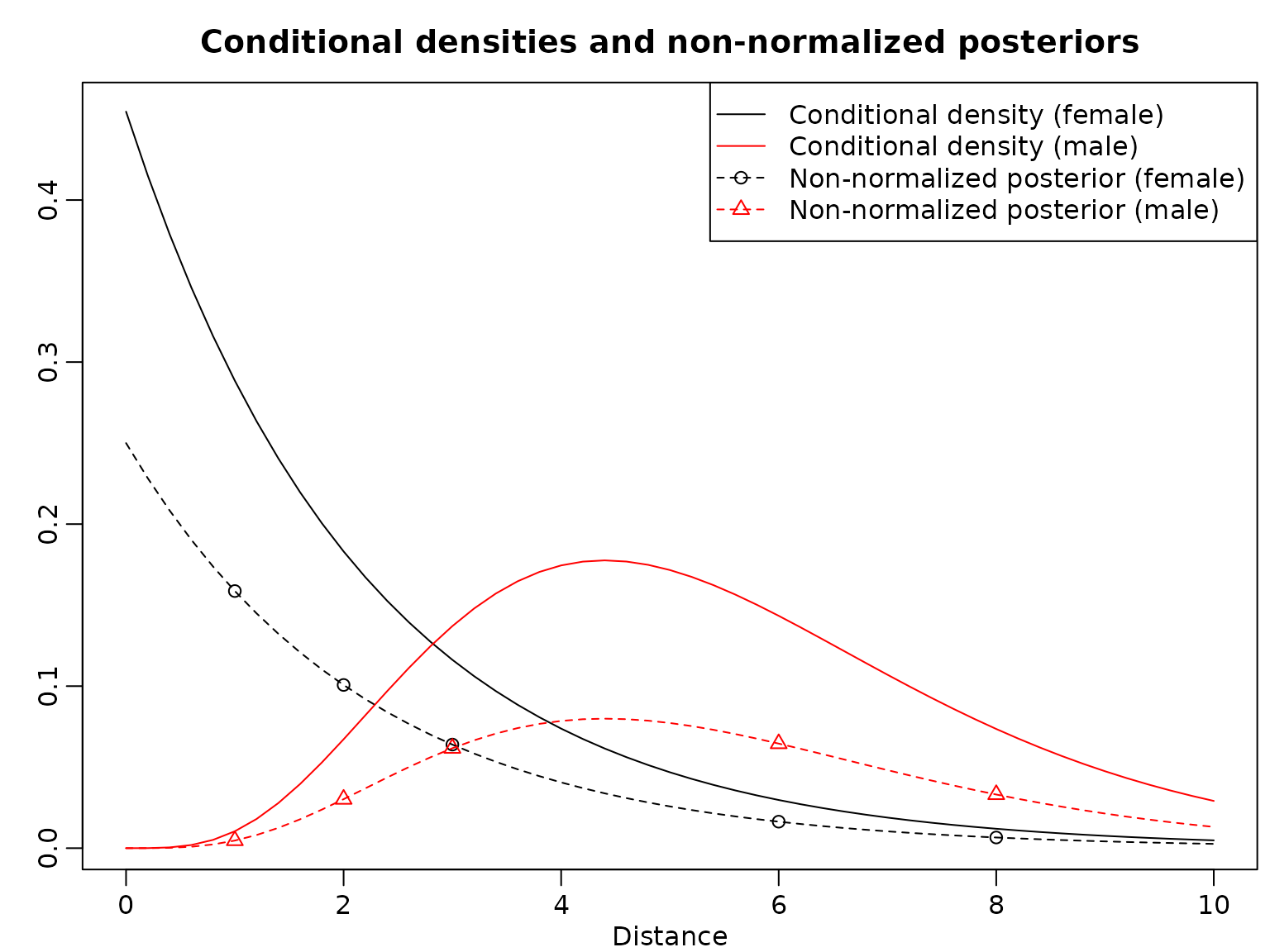
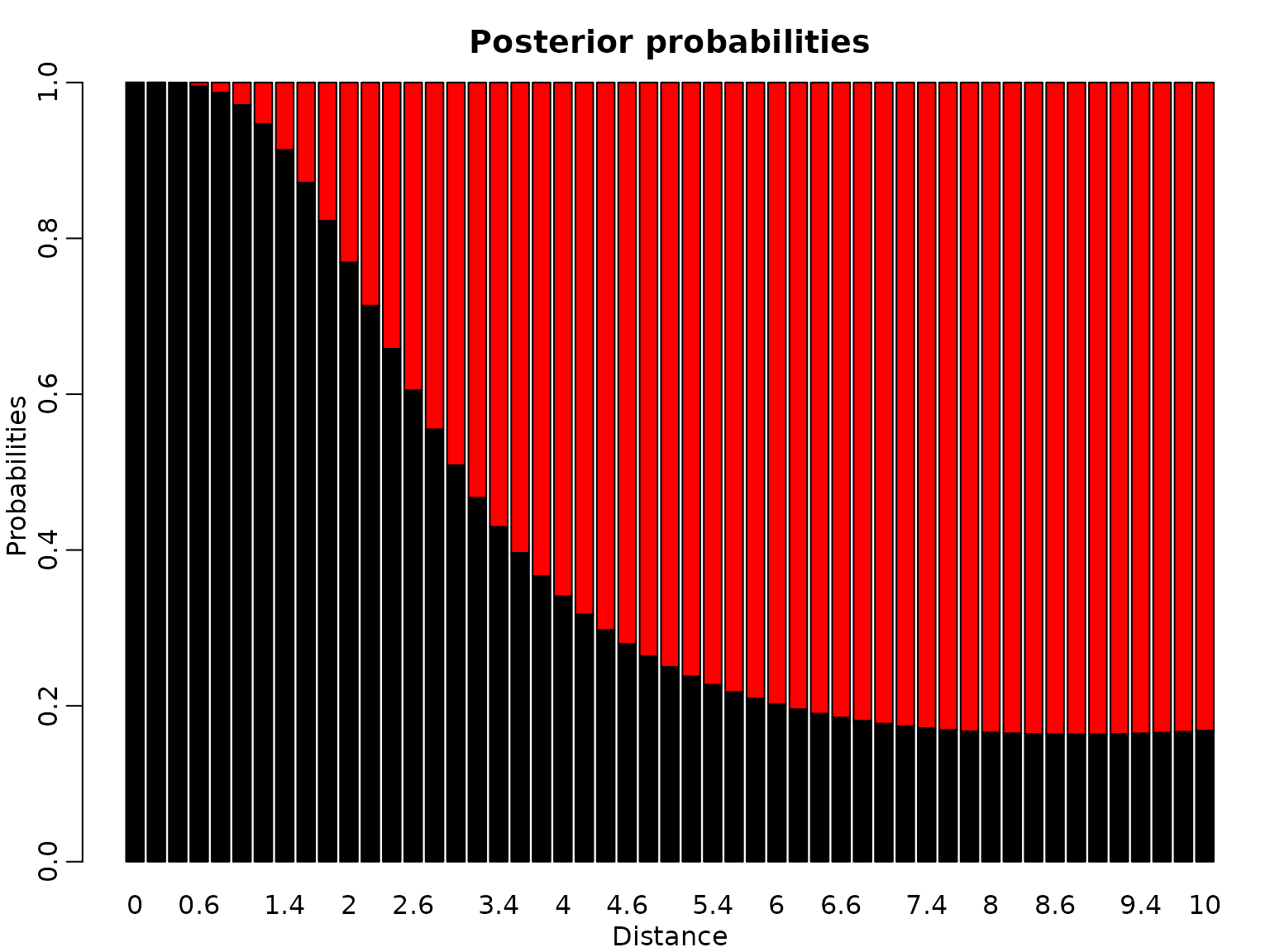
Finally, here are the posterior probabilities. Note that each ratio of the pairs of corresponding posterior probabilities is the same as each ratio of the heights of the vertically corresponding points in the graph.
normalizer <- theta1_y_unnormalized + theta0_y_unnormalized
theta1_y <- theta1_y_unnormalized / normalizer
theta0_y <- theta0_y_unnormalized / normalizer
res <- rbind(female = theta1_y, male = theta0_y)
colnames(res) <- y
knitr::kable(round(res, 3))| 1 | 2 | 3 | 6 | 8 | |
|---|---|---|---|---|---|
| female | 0.971 | 0.769 | 0.509 | 0.202 | 0.166 |
| male | 0.029 | 0.231 | 0.491 | 0.798 | 0.834 |
We also plot the posterior probabilities for distance values ranging from 0 to 10 in steps of 0.2 in a stacked barplot.
normalizer <- theta1_y_all_unnormalized + theta0_y_all_unnormalized
theta1_y_all <- theta1_y_all_unnormalized / normalizer
theta0_y_all <- theta0_y_all_unnormalized / normalizer
res <- rbind(female = theta1_y_all, male = theta0_y_all)
colnames(res) <- y_all
barplot(res, main = "Posterior probabilities", xlab = "Distance",
ylab = "Probabilities", col = c("black", "red"))